| Michael W. Hunkapiller |
 |
|
 |
 |
 |
|

[figure 39]

[figure 40]

[figure 41]
[figure 42]

[figure 43]
[figure 44]

[figure 45]

[figure 46]
[figure 47]
[figure 48] |
One of the
commercial initiatives that the partnership between Applied Biosystems,
Celera Genomics, and our newly created joint venture, Celera Diagnostics,
has added is an initiative to look at diversity across the human
genome. [figure 39] The key, here, is to be able to re-sequence
all of the gene areas within the chromosomes and create assays that
allow individual researchers to study whichever portion of the genome
they want. [figure 40] For example, to look at the variation of
cells within that sequence, to look at the expression of those individual
genes in a robust way, and to be able to look at all the sites of
diversity, it is useful for researchers to not have to start that
process to create their own assays, but to have ones that are already
validated by a commercial company.
The final area I will touch on is proteomics, an area which I know
has a great deal of importance in some of the strategic, long-term
research plans within Japan, as elsewhere in the world. [figure
41] In this respect, Applied Biosystems and the tools that we developed
initially at Lee Hood's lab have played a role already. For instance,
the first small-scale protein sequencer was developed at Lee's lab
and was the first commercial product provided by Applied Biosystems
in 1982. [figure 42] This protein sequencer was used, particularly
by biotech companies, to look at key, biologically functional proteins.
The sequencer aided these companies in the process to eventually
commercialize some of the key proteins as pharmaceutical compounds.
[figure 43] The problem of looking at not just a single protein,
but the proteome-wide collection of proteins, is a daunting one.
One doesn't have a PCR tool that allows you to make unlimited quantities
of individual proteins, so you have to rely on what's there -- which
might not be very much in a lot of cases. The number of proteins
that exist and the complexity of their chemical make-up are much
greater than in the case of DNA.
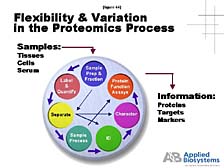
One needs to understand that it's a complicated process. It starts
with the fact that the way you prepare a particular protein for
analysis is going to be different from the way that you prepare
another one. [figure 44] Whereas, in the DNA world, it's pretty
much the same. You are also after a much more complicated set of
information in the case of proteins. Therefore, there is not likely
to be a single tool that does what DNA sequencing, or even PCR,
has done for nucleic acid studies. Instead, there are some tools
that allow you to attack specific areas of protein structure and
function problems. [figure 45] One of these that we've focused on,
and what Craig will touch on a little bit more, is mass spectrometry.
It is an old tool, from a chemical analysis perspective, and one
that's increasingly applied to protein studies. The difficulty,
here, is there are a lot of bottlenecks. It's hard to prepare a
large number of samples for analysis for very large-scale proteomic
studies. It's even more difficult to interpret the data that one
generates by analytical processes such as mass spectrometry. Thus,
the process doesn't have nearly as high a throughput as what's been
achieved for DNA analysis in the last few years.
The goal that we have now in looking at new technologies is to eliminate
those bottlenecks, so one can have a high throughput in proteomics-level
studies. This is really where functional activity occurs within
a cell. It is a challenge which has not been fully realized yet,
but one in which there is a substantial amount of effort, from our
own laboratories, and others as well.
One of the tools that we have recently announced is a new generation
of mass spectrometry tools applied specifically to the proteomics
challenge. [figure 46] This tool allows you to cut down, to some
degree, on sample prep problems because they can analyze accurately
more complicated protein mixtures, and do it in a much faster rate
for components than what was possible with the earlier technology.
Therefore, we hope to present this as the first "proteomics analyzer,"
at least for structural proteomics.
That said, hard work can solve a lot of the problems, for example,
work in areas such as the chemistry required to be able to prepare
the elements of proteins that you want to study. Remember the study
of very complicated molecules is in its infancy as well. One of
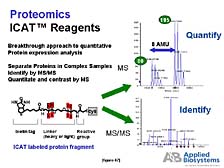
the newer tools, developed by Reudi Aebersold at the University
of Washington in Seattle, is one that we have been commercializing
in order to help address some of these problems. [figure 47] This
tool allows differential isotope-labeling methodologies to look
at protein expression, which has been a particularly intractable
problem in the case of proteins for which you don't have a good,
very specific functional activity assay. While we have commercialized
this technology only recently, some of the large proteomics labs,
such as Celera and Oxford Glycosystems in the U.K., are already
reporting fairly remarkable results with its use to study protein
expression in a broad sense.
Let me end my presentation by reminding you where I started. We
are in the business of providing tools that enable the life sciences
research community to carry out the basic task of understanding
the function of biological systems. This includes finding out how
one can intervene when there are problems in the form of medical
therapies, how one can help diagnose problems, and how one can use
biology as a tool to solve a host of commercial problems, from agriculture
to the identification of criminals. [figure 48] The bottom line
is that no single discipline can solve this problem. Biology is
a complex enough endeavor that it requires the combined effort and
expertise of many disciplines to produce analytical systems that
can help unravel these problems. We are proud that we have been
a part of helping spur this process over the last twenty years or
so, and we would expect to continue to do so going forward into
the future.
With that I will turn it back over to Dr. Matsubara.
MODERATOR: Thank you very much, Dr. Hunkapiller, for a wonderful
presentation. |
|
|